Fundamental Science
Research Direction 2:
Role of FLT3 Signaling in the Brain
A variety of brain diseases manifest epileptic seizures or subconvulsive epileptiform discharges, either as a main symptom or as a comorbidity. Therefore, the imbalance between excitation and inhibition (E/I balance) in brain circuits may point to a common disease mechanism that underlies these diseases. Our previous work used a human patient-derived neuron model of Rett syndrome (RTT), a severe neurodevelopmental disorder, to make the discovery that reduced expression of a key neuronal gene that regulates E/I balance, KCC2, causes the functional abnormalities observed in RTT neurons (Tang et al., PNAS, 2016). We further validated our discovery of KCC2 expression reduction in a Mecp2 mutant animal model of Rett syndrome, and demonstrated that such deficiency contributes to neural circuit function abnormalities (Banerjee et al., PNAS, 2016).
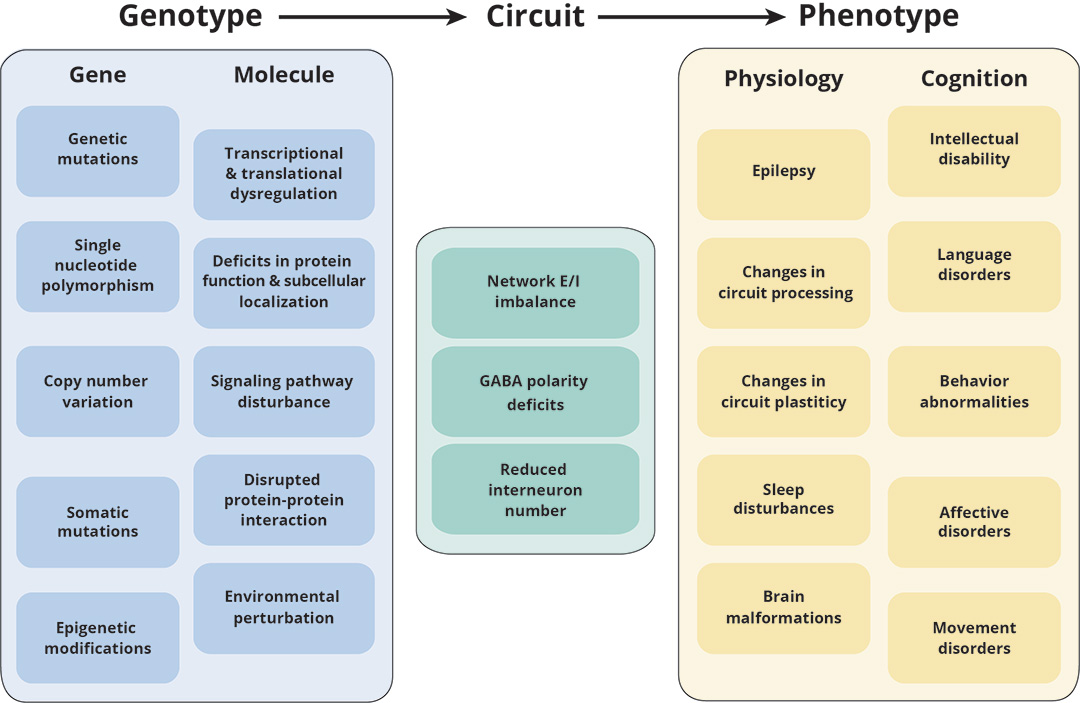
Pathogenic mechanisms underlying neurodevelopmental disorders (Tang et al., Nature Reviews Neuroscience, 2021)
From our unbiased high-throughput screening to identify compounds that enhance KCC2 gene expression, we unexpectedly discovered a number of FLT3 kinase pathway inhibitor drugs, which are approved by the FDA to treat blood cancer but have not been well studied in the brain. Our mRNA sequencing results uncovered previously unknown roles of the FLT3 kinase pathway in promoting synapse development and reducing neuroinflammation. We are currently employing a combination of research techniques including genome-engineered human cells and mouse models, RNA sequencing, and genome-wide CRISPR screening, and electrophysiology, to elucidate the potentially diverse roles of the FLT3 pathway in modulating signal transduction and gene expression in brain cell types. We will further map the mechanistic insights gained from these discoveries onto brain function and dysfunction to drive the development of therapeutics.
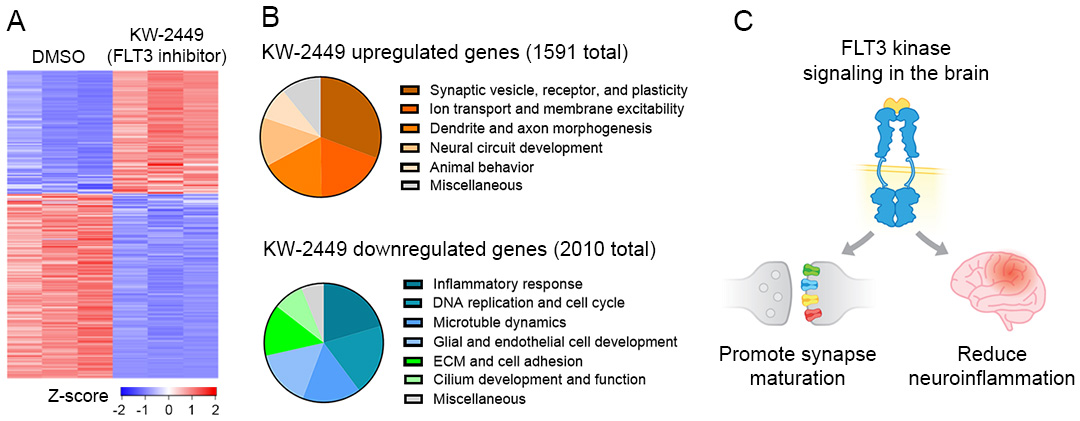
Inhibiting FLT3 signaling induces selective changes in brain gene expression. (unpublished result, Tang lab, 2021)
Related Publications
1. Tang, X., Sur, M., Jaenisch, R.. The role of GABAergic signalling in neurodevelopmental disorders, Nature Reviews Neuroscience, PMID: 33772226
2. Tang, X., Kim, J., Zhou, L., Wengert, E., Zhang, L., Wu, Z., Carromeu, C., Muotri, AR., Marchetto, MC., Gage, FH., Chen, G.. KCC2 rescues functional deficits in human neurons derived from patients with Rett syndrome, Proceedings of the National Academy of Sciences, 2016. PMCID: PMC4725523. (highlighted by F1000Prime)
3. Banerjee, A., Rikhye, RV., Breton-Provencher, V., Tang, X., Li, C., Li, K., Runyan, CA., Fu, Z., Jaenisch, R., Sur, M.. Jointly reduced inhibition and excitation underlies circuit-wide changes in adult cortical processing in vivo, Proceedings of the National Academy of Sciences, 2016. PMCID: PMC5135376.
4. Tang, X. (chief editor), Neuronal Chloride Transporters in Health and Disease, Academic Press, 2020, paperback ISBN: 9780128153185, eBook ISBN: 9780128153192 (A 26-chapter framework for this book to highlight the recent advances in the basic biology and disease-related research in the neuronal chloride transporter field, recruited and worked with over 80 authors who are leading experts in their respective subfields to accomplish this 766-page book, which was published in July 2020.)
5. Tang, X., Zhou, L., Wagner, AM., Marchetto, MC., Muotri, AR., Gage, FH., Chen, G.. Astroglial cells regulate the developmental timeline of human neurons differentiated from induced pluripotent stem cells. Stem Cell Research, 2013, PMCID: PMC397996. (cover story)
6. Tang, X., Genome Editing: Applications for Disease Modeling and Cell Therapy, Wiley publishing company, 2019.


